Biological Chemistry
- B.A. Columbia College (1962)
- Ph.D. Harvard University (1968)
- NATO postdoctoral fellow
- Institut Pastuer, Paris (1967-68)
- Merck Faculty Award (1970)
- Sloan Foundation Fellow (1974-77)
- N.I.H. Physical Biochemistry Study Section (1978-82)
- Vice Provost for Research (1982-1995)
- Chevalier de l’Ordre des Palmes Académique (2004)
- Fellow of the American Association for the Advancement of Science (2004)
Our overall thrust is to study the linkage between biological structure and function, using a broad array of chemical, physical, and biological tools. Our major efforts fall in three principal areas.
Ribosomes
We are interested in the structure and function of the bacterial ribosome, which is the site of protein biosynthesis in the cell. Most recently we have been focusing on the application of kinetic and spectroscopic approaches, including the use of single molecule and single-turnover studies in conjunction with fluorescence resonance energy transfer (FRET), to elucidate mechanisms for ribosomal catalysis of protein synthesis. We are particularly interested in the functions of G-proteins on the ribosome, and how these functions are altered by antibiotics and by mutations of tRNAs and ribosomal RNA. We are also pursuing studies on how the rate of protein synthesis is modulated by specific mRNA and oligopeptide sequences.
Ribonucleotide reductase (RR)
RR catalyzes the reduction of nucleoside diphosphates to deoxynucleoside diphosphates and is the key enzyme controlling the rate of DNA synthesis. As such it is highly regulated and is a target enzyme for cancer chemotherapy. Our studies focus on the RRs derived from mammalian cells. We are developing novel and specific inhibitors of this enzyme, using both a rational design approach and combinatorial methods, and based in part on our detailed studies of the allosteric regulation of this enzyme. This work utilizes synthetic organic chemistry, biochemistry, molecular modeling and X-ray crystallography approaches.
Serine proteinase inhibitors ("serpins")
Serpins are known to be of great importance for inflammation process in mammals. We seek to understand the structural basis for the specificity of interaction of these serpins with a variety of serine proteases, using a combination of chemical modification, single turnover FRET kinetics, FT-IR spectroscopy and genetic engineering approaches to elucidate the basic mechanisms underlying such specificity. A second area of interest is serpin polymerization, which underlies several diseases associated with serpin malfunction. These studies are being carried out using single molecule confocal microscopy.
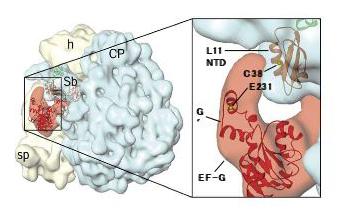
Dynamics of Ribosomal Conformational Change
Wang Y, Qin H, Kudaravalli RD, Kirillov SV, Dempsey GT, Pan D, Cooperman BS, Goldman YE (2007) Single Molecule Structural Dynamics of EF-G·Ribosome Interaction During Translocation, Biochemistry. in press
Grigoriadou C, Marzi S, Pan D, Gualerzi CO, Cooperman BS (2007) The Translational Fidelity Function of IF3 During the Transition from 30S to 70S Initiation Complex. J. Mol. Biol., in press, doi:10.1016/j.jmb.2007.07.031
Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS (2007) A Quantitative Kinetic Scheme for 70S Translation Initiation Complex Formation. J. Mol. Biol., in press,
doi:10.1016/j.jmb.2007.07.032
Chowdhury P, Wang W, Bunagan MR, Klemke JW, Tang J, Lavender S, Saven JG, Cooperman BS, Gai F (2007) Fluorescence Correlation Spectroscopic Study of Serpin Depolymerization by Computationally Designed Peptides. J Mol Biol. 369, 462-73.
Pan D, Kirillov S, Cooperman BS (2007) Kinetically Competent Intermediate(s) in the Translocation Step of Protein Synthesis. Molecular Cell,. 25, 519-529.
Pan D, Kirillov S, Zhang CM, Hou YM, Cooperman BS (2006) Rapid Ribosomal Translocation Depends on the Conserved 18:55 Base Pair in P-site tRNA. Nature Structural and Molecular Biology, 13, 354-9.
Seo HS, Abedin S, Kamp D, Wilson DN, Nierhaus KH, Cooperman BS (2006) EF-G Dependent GTPase on the Ribosome. Conformational Change and Fusidic Acid Inhibition. Biochemistry 45, 2504-14.
He J, Roy B, Perigaud C, Kashlan OB, Cooperman BS (2005) The enantioselectivities of the active and allosteric sites of mammalian ribonucleotide reductase. FEBS J. 272,1236-42.
Gao Y, Kashlan OB, Kaur J, Tan C, Cooperman BS (2005) Mechanisms of action of peptide inhibitors of mammalian ribonucleotide reductase targeting quaternary structure. Biopolymers (Peptide Science), 80, 9-17.
Purkayastha P, Klemke JW, Lavender S, Oyola R Cooperman BS, Gai F (2005) α1-Antitrypsin polymerization: A fluorescence correlation spectroscopic study. Biochemistry, 44, 2642-2649.
Seo HS, Kiel M, Pan D, Raj VS, Kaji A Cooperman BS (2004) Kinetics and Thermodynamics of RRF, EF-G, and Thiostrepton Interaction on the E. coli Ribosome. Biochemistry 43, 12728-40.
Tan C, Gao Y, Kaur J, Kashlan, O. B., Cooperman BS (2004) More potent linear peptide inhibitors of mammalian ribonucleotide reductase. Bioorg. Med. Chem. Lett., 14, 5301-5304.
Kashlan OB, Cooperman BS (2003) Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 42(6): 1696-1706.
Gao Y, Liehr S, Cooperman BS (2002) Affinity-Driven Selection of Tripeptide Inhibitors of Ribonucleotide Reductase. Bioorg. Med. Chem. Lett., 12, 513-515.
Hsieh MC, Cooperman BS (2002) The Inhibition of Prostate-Specific Antigen (PSA) by a-Antichymotrypsin: Salt-Dependent Activation Mediated by a Conformational Change. Biochemistry 41, 2990-2997.
Kashlan OB, Scott CP, Lear JD, Cooperman BS (2002) A Comprehensive Model for the Allosteric Regulation of Mammalian Ribonucleotide Reductase. Functional Consequences of ATP- and dATP-Induced Oligomerization of the Large Subunit. Biochemistry 41, 462-474.
Scott CP, Kashlan OB, Lear JD, Cooperman BS (2001) A Quantitative Model for Allosteric Control of Purine Reduction by Murine Ribonucleotide Reductase. Biochemistry 40, 1651-1661.
O’Malley KM, Cooperman BS (2001) Formation of the covalent chymotrypsin: antichymotrypsin complex involves no large-scale movement of the enzyme. J. Biol. Chem., 276, 6631-6637.
Pender BA, Wu X, Axelsen PH, Cooperman BS (2001) Toward a Rational Design of Peptide Inhibitors of Ribonucleotide Reductase: Structure - Function and Modeling Studies. J. Med. Chem., 44, 36-46.

